Comprehensive Analysis of Aerobic and Anaerobic Bacteria Found on Dental Bib Clips at a Hygiene Clinic
Abstract
Multiple-use dental bib clips are considered to present relatively low risks for transmitting infections and, thus, are thought to only require disinfection between patient visits. This study was designed to: 1) determine the presence and composition of bacterial contaminants on reusable rubber-faced metal bib clips after dental treatment at the hygiene clinic at Tufts University School of Dental Medicine and 2) evaluate the effectiveness of the disinfection for this clip type. Aerobic and anaerobic bacterial contaminant loads on the surfaces of the clips were investigated immediately after hygiene treatments were rendered and again after clips were disinfected. The species and strains of bacterial isolates were identified using 16S rDNA sequencing and Human Oral Microbe Identification Microarray analyses. The results demonstrated that although the use of disinfection proved to be significantly effective, some clips retained at least one bacterium on their surfaces after disinfection. Although the bacterial species present on disinfected clips were typical skin or environmental isolates, some were oral in origin. In the study’s settings, bacterial presence on the clips did not indicate an infectious disease problem. The different bacterial loads on clips suggest that cross-contamination risks may not be the same for all clinics, and that this difference may be related to the type of treatments and services performed.
Medical devices, instruments, and materials in the dental office are either designed to be discarded immediately after each patient visit or are meant to be reused for multiple patient visits. Bib chains, dental mirrors, and ultrasonic and sonic scaling tips are examples of reusable items. Reprocessing and reusing these products require adequate disinfection or sterilization processes, which are performed to prevent or significantly reduce the risk for cross-contamination and disease transmission between the patient and dental healthcare provider, as well as among patients. According to the Centers for Disease Control and Prevention guidelines, items contacting only intact skin, such as bib chains, have a low risk for transmitting infection. The guidelines suggest cleaning and disinfection of these items with an intermediate- or low-level hospital disinfecting product registered with the Environmental Protection Agency.1
Molinari2 showed that bacteria can be found on chains after dental treatments are rendered and that a quick cleaning of these chains with intermediate-level, environmental surface disinfectant wipes between patient treatments did not completely eliminate the presence of bacterial contaminants. Chains that were reused throughout the day without being cleaned between patients’ appointments showed high concentrations of bacteria on their surfaces.2 A study performed at the Dental School of Witten/Herdecke University in Germany also showed the presence of bacterial microorganisms on clips after both dental treatment and disinfection.3 A separate study by the University of North Carolina at Chapel Hill’s School of Dentistry revealed similar results, in addition to the finding of pathogenic bacteria on the clips.4
Our previous research at clinics that specialize in endodontic or orthodontic treatments demonstrated that bib clips had significantly different loads of aerobic bacteria on their surfaces immediately after patient care. After disinfection, 70% to 80% of the sampled clips did not show any aerobic bacterial contaminants; nonetheless, 20% to 30% of the disinfected clips retained some aerobic bacterial contaminants on their surfaces.5 The results of that study indicated the levels of risk for cross-contamination differed depending on the nature of the dental treatment.5 In the present study, we identified after treatment and again after disinfection the aerobic and anaerobic bacterial contaminants on the surfaces of reusable, rubber-faced metal clips in a clinic that focuses on hygiene treatments.
Objective
The primary objectives were to: 1) determine the presence and composition of bacterial contaminants on clips of reusable patient bib chains after dental treatment at the hygiene clinic, and 2) evaluate the effectiveness of disinfection for these clips using a commercially available disinfectant.
Materials and Methods
Clips and Tested Parameters
Reusable, rubber-faced metal clips that were attached to rubber chains (Slip-Nots™, Kerr TotalCare, Metrex Research Corp., www.metrex.com) were used in the hygiene clinic at Tufts University School of Dental Medicine (TUSDM) for the purpose of this study. A total of 20 chains were analyzed; from each chain, only the clips were sampled for the presence of aerobic and anaerobic bacterial contaminants. The study protocol was approved by Tufts Medical Center and Tufts University Health Sciences Institutional Review Board Office.
Bacterial Sample Collection
Bib clips were sampled for bacterial contaminants immediately after hygiene treatments were provided to patients at the hygiene clinic. Samplings were performed immediately after patient treatment (post-treatment clips) and again after the clips were disinfected (post-disinfection clips) using CaviCide® disinfecting wipes (CaviWipes®, Metrex Research Corp.) and drying them according to the manufacturer’s instructions. The disinfection protocol followed at TUSDM was applied. The active ingredients in these wipes are 17.2% isopropanol and 0.28% diisobutylphenoxyethoxyethyl dimethyl benzyl ammonium chloride. Immediately after each patient discharge, the surfaces of each clip were sampled in the operatory for the presence of aerobic and anaerobic bacteria. To ensure randomization, the statistical software package R version 2.11.1 (cran.r-project.org) was used to help assign the clips from each chain to aerobic or anaerobic bacteria samplings, prior to the study commencement. For each chain, bacterial contaminants were directly transferred from one clip onto a Trypticase soy agar plate for the growth of aerobic bacteria (Trypticase™ Soy Agar TSA II, BD, www.bd.com). Bacterial contaminants were directly transferred from the opposing clip and onto a 5% sheep blood agar plate (BBL™ Trypticase™ Soy Agar with 5% Sheep Blood [TSA II], BD) to test for the growth of anaerobic bacteria. A direct transfer was achieved by firmly pressing each of the four sides of every clip onto a separate area on a nutrient agar plate, and this process was repeated several times across the agar surface.6
Thereafter, both clips were disinfected with CaviCide disinfecting wipes and, using the same method, were sampled again for bacterial contaminants on separate agar plates. To generate and maintain an anaerobic growth environment, the 5% sheep blood agar plates were placed in a GasPak™ EZ Anaerobe Pouch System (BD) in the operatory. Plates were then transferred to a microbiology laboratory at TUSDM. Trypticase Soy Agar plates (BD) were incubated in ambient atmosphere at 37°C for 48 hours, and the 5% sheep blood agar plates were incubated at 37°C in anaerobic conditions for 6 days. For each plate, the number of bacterial colonies that grew on the agar was recorded and representative plates were imaged.
Bacterial Identification
Genetic identification of bacterial colonies that developed on the Trypticase Soy Agar plates and 5% sheep blood agar plates was performed at the Human Microbial Identification Microarray Core at The Forsyth Institute (Cambridge, MA; mim.forsyth.org). The species and strains of colonies that developed on the nutrient agar plates from post-disinfection clips were individually identified using 16S rDNA sequencing analysis. Human Oral Microbe Identification Microarray (HOMIM) analysis was used to identify the species and strains of multiple bacterial colonies that developed from post-treatment clips.7,8 In a single hybridization reaction, this innovative, high-throughput technology enables researchers to identify approximately 300 common oral bacterial species, including not-yet-cultivated phylotypes, from a given clinical sample.8-10
Sample Size Calculation
A sample size calculation was conducted using the statistical software package R version 2.11.1. Assuming that 100% of the clips would not harbor pathogenic bacteria such as Escherichia coli on their surfaces following hygiene treatments, a sample size of 20 was adequate to obtain a 95% confidence interval between 86% and 100%.
Statistical Analysis
The authors used means, medians, and upper and lower quartiles, as well as minimum and maximum numbers of bacterial colonies, to analyze the findings from sampling the surfaces of post-treatment and post-disinfection clips. Nonparametric hypothesis testing was conducted using the Wilcoxon signed-rank test because the data showed non-normality. P values lower than .05 were considered statistically significant. All descriptive statistics and hypotheses tests were completed using SAS software, version 9.2 (SAS Institute Inc., www.sas.com). Exact 95% confidence intervals for the percentage of clips that did not exhibit any non-oral pathogens were calculated using binomial distribution.
Results
Bacterial Load on Post-Treatment Clips
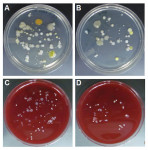
In keeping with findings from previous studies,3,4 the present authors found that aerobic and anaerobic bacterial contaminants can be found on clips that were sampled immediately following a hygiene treatment (Table 1). A total of 1,640 bacterial colonies grew on the Trypticase soy agar plates in aerobic conditions, and 2,405 bacterial colonies grew on the 5% sheep blood agar plates in anaerobic conditions (Table 1, A). Per sampled post-treatment clips from each chain, an average of 82.0 and 120.3 bacterial isolates grew in aerobic and anaerobic conditions, respectively (Table 1, B). When stratified by growth conditions, the total bacterial amount found on post-treatment clips revealed a 1.5-fold increase in the counts of anaerobic bacterial colonies in comparison with those of the aerobic bacterial colonies (Table 1). The descriptive statistics of bacterial colony counts, stratified by clips tested and then by bacterial growth conditions, are shown in Table 2, A. The data demonstrate that after dental hygiene treatment, no statistically significant difference between the counts of anaerobic and aerobic bacterial isolates from samples of post-treatment clips (P = .084) was shown (Table 2, A). Figure 1 demonstrates representative images of the growth of aerobic bacterial colonies on Trypticase soy agar plates (Figure 1, A and B) and anaerobic bacterial colonies on 5% sheep blood agar plates (Figure 1, C and D) from clips sampled following hygiene treatment.
Identification of Bacterial Isolates Sampled from Post-Treatment Clips
The species and strains of the bacterial colonies that grew aerobically and anaerobically were identified using 16S rDNA sequencing and HOMIM analyses at the Human Microbial Identification Microarray core.7,8 As demonstrated in Table 3, A, the predominant bacterial isolates collected from the surfaces of the 20 post-treatment clips and developed under ambient aerobic conditions were identified as Staphylococcus caprae and/or Staphylococcus epidermidis (19 of 20 clips), Synergistetes [G-3] sp (14 of 20 clips), Slackia exigua (8 of 20 clips), and Prevotella oris or Prevotella cluster IV, and Streptococcus cluster II (6 of 20 clips), as well as Streptococcus oralis, Streptococcus parasanguis I, Streptococcus parasanguis II, Pseudomonas cluster I, and Neisseria cluster II (5 of 20 clips). In anaerobic growth conditions, both skin and oral bacteria were found on the clips. The major bacterial isolates that developed were identified as S caprae and S epidermidis (19 of 20 clips); Streptococcus cluster II (18 of 20 clips); Staphylococcus anginosus, Staphylococcus intermedius, and Actinomyces georgiae (17 of 20 clips); S exigua (16 of 20 clips); S oralis (14 of 20 clips); Propionibacterium acnes and P oris, Streptococcus sanguinis (13 of 20 clips); Streptococcus anginosus and Streptococcus gordonii (12 of 20 clips); Haemophilus parainfluenzae, S parasanguis I, S parasanguis II, and Staphylococcus salivarius (11 of 20 clips); Parvimonas micra (10 of 20 clips); Capnocytophaga granulosa, Neisseria cluster II, and Veillonella parvula and/or Veillonella dispar (9 of 20 clips), Streptococcus australis, Fusobacterium nucleatum ss nucleatum, and/or Fusobacterium nucleatum ss animalis, and Synergistetes [G-3] sp (8 of 20 clips), Eikenella corrodens, Kingella denitrificans, Streptococcus infantis, Streptococcus parasanguinis I, and Streptococcus parasanguinis II (7 of 20 clips), Haemophilus sp, Campylobacter gracilis, Gemella morbillorum, Streptococcus anginosus, Streptococcus intermedius, Streptococcus cristatus, Streptococcus constellatus, and Synergistetes sp (6 of 20 clips), and Prevotella cluster IV, Streptococcus cluster I, Leptotrichia buccalis, Leptotrichia goodfellowii, and Sneathia sanguinegens (5 of 20 clips). In addition, species and strains of aerobic, as well as anaerobic bacterial, isolates appeared less commonly and these bacteria were sporadically isolated from individually sampled clips (Table 3).
Bacterial Load on Post-Disinfection Clips
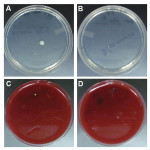
After disinfection of the clips and when clips were sampled for the presence of aerobic and anaerobic bacterial contaminants, the amount of bacterial colonies that developed on the nutrient agar plates markedly decreased (Table 1). A total of 23 aerobic bacterial colonies and 72 anaerobic bacterial colonies grew on Trypticase soy agar plates and 5% sheep blood agar plates, respectively (Table 1, A). After disinfection, with the total number of sampled clips taken into account, an average of 1.2 to 3.6 bacterial isolates grew from the aerobic and anaerobic conditions, respectively, per chain (Table 1, B). Figure 2 shows representative images of aerobic bacterial colonies growth on Trypticase soy agar plates (Figure 2, A and B) and of anaerobic bacterial colonies growth on 5% sheep blood agar plates (Figure 2, C and D) from the surfaces of disinfected clips. As shown in Table 2, A, the descriptive statistics of bacterial colonies counts, stratified by clips tested and then by bacterial growth conditions, did not demonstrate a statistically significant difference when comparing the total counts of aerobic and anaerobic bacterial isolates in the post-disinfection setting (P = .0514). When stratified by growth conditions and then by clips tested, there was a statistically significant difference between the post-treatment and post-disinfection settings, both under aerobic (P < .0001) as well as anaerobic (P < .0001) growth conditions (Table 2, B). Finally, the results in Table 1, C indicate that at least one aerobic bacterial colony developed after sampling the surfaces of clips from eight chains, representing 40% of the tested chains. Similarly, one or more anaerobic bacterial colonies developed after sampling the surfaces of clips from 14 chains, representing 70% of the tested chains. Similar to findings from previous studies,2-5 these data show that aerobic and anaerobic bacteria can be detected on clips following disinfection.
Identification of Bacterial Isolates from Post-Disinfection Clips
Table 4 demonstrates that S epidermidis was the major bacterial isolate collected from the surfaces of 20 post-disinfection clips and developed under aerobic conditions (6 of 20 clips). Under these conditions, Staphylococcus hominis, Neisseria flava, Streptococcus salivarius, and different species of Bacillus were also found sporadically on single clips. The main bacterial isolates that developed under anaerobic conditions were identified as P acnes (9 of 20 clips), S epidermidis (4 of 20 clips), and Streptococcus mitis (3 of 20 clips). In addition, S hominis, Streptococcus constellatus, S salivarius, and S sanguinis, Prevotella dentalis, Neisseria flavescens, Rothia mucilaginosa, Kocuria marina, Veillonella dispar, Actinomyces sp, Dermabacter hominis, and Selenomonas noxia appeared less commonly and were isolated from individual clips. In contrast to findings from a previous study,4 after treatment and disinfection, the surfaces of all clips tested at the hygiene clinic at TUSDM had no non-oral pathogenic bacteria, such as E coli. Based on these results, we can state with 95% confidence level that the percentage of chains that would show no non-oral pathogenic bacteria under the tested conditions would be 86%-100%.
Discussion
The study was performed in a hygiene clinic where the authors identified aerobic and anaerobic bacterial species that were present on the surfaces of reusable, rubber-faced metal clips immediately following dental treatments and after disinfection of these clips. Typically, patients are referred to this hygiene clinic for periodontal maintenance therapy after they have completed active periodontal treatments. In addition, individuals with no history of periodontal diseases visit this clinic for prophylaxis or further referrals. Hygiene services performed at this clinic include intra- and extraoral examinations, gingival assessments, radiographs, fluoride treatments, and prophylaxis with the use of both hand instruments and ultrasonic scalers.
Similar to findings reported in previous studies,2-5 the present results showed that the surfaces of sampled clips contained bacterial contaminants at the conclusion of a dental treatment. However, the settings for this study allowed us to specifically investigate the bacterial loads on clips in a clinic that specializes in hygiene treatments, rather than on clips collected simultaneously from multiple clinics with different specialties.2-4 Our previous study, published in Compendium of Continuing Education in Dentistry (www.dentalaegis.com/special-issues/Dux/2012/08/), at the orthodontics and endodontics clinics at TUSDM demonstrated that 20% and 25% of these rubber-faced metal clips retained few aerobic bacterial contaminants on their surfaces after disinfection, respectively.5 Although the results of the present study found 60% of the clips had no evident aerobic bacterial contaminants after disinfection, the results also showed that 40% of the clips had at least one type of aerobic bacteria. Based on the results from our previous and present studies, it is likely that the type of dental treatment affects the levels of bacterial loads on clips before and after disinfection. However, it remains unclear if, in addition to different bacterial loads on clips, bacterial contaminants are different in their species among dental clinics with different specialties. If so, both the presence and composition of bacterial contaminants on these clips may indicate that various clinics are exposed to different contamination risks. In addition, the importance of incubation of bacterial contaminants from the surfaces of clips in both aerobic and anaerobic optimal growth conditions was demonstrated in the present study. The authors found that although 40% of post-disinfection, rubber-faced metal clips had at least one type of aerobic bacterium on their surfaces, 70% of these clips revealed the presence of few anaerobic bacteria on their surfaces. Nonetheless, in both cases, the data showed a statistically significant difference when bacterial loads on post-treatment and post-disinfection clips were compared, indicating that the disinfection procedure was significantly effective. These findings raise the question as to why aerobic bacteria, and specifically fastidious anaerobic bacteria, can persist on the surfaces of clips 3 minutes after disinfection. One possibility is that the time that the bacteria is exposed to the disinfectant may not be sufficient to eliminate all bacteria. This would suggest that more time should be allowed between disinfecting the clips and sampling the bacteria; to test this possibility, kill curves of bacterial survival should be performed. Alternatively, the effect of the disinfection procedure on specific aerobic and anaerobic bacteria may be bacteriostatic rather than bactericidal. Thus, in supportive optimal growth conditions, remaining viable bacteria can continue developing.
Patients and dental healthcare providers, such as dentists, hygienists, and assistants, are likely to be exposed to normal oral flora or pathogenic microorganisms that inhabit or infect the oral cavity and respiratory tract.1 Using 16S rDNA sequencing and HOMIM analyses,7,8 we identified the species and strains of the bacterial isolates on the surfaces of the tested clips. We found that immediately after treatment, in both aerobic and anaerobic growth conditions, the predominant bacterial isolates on the surfaces of 95% of post-treatment clips were Staphylococcus epidermidis and S caprae, which are the most common bacterial flora that reside on skin and mucous membranes.11 In addition, in aerobic growth conditions, strains of Synergistetes and Slackia were found on 70% and 40% of the clips, respectively. Under anaerobic conditions, bacterial isolates included strains of Streptococcus, Actinomyces, Slackia, Propionibacterium, Prevotella, Haemophilus, Parvimonas on the surfaces of 50% to 90% of the clips. Other species of environmental and oral bacteria were also found randomly on post-treatment clips in aerobic and anaerobic conditions. After disinfection, we found that S epidermidis was the main bacterial isolate from 30% of the clips under aerobic growth conditions, while P acnes was the major bacteria isolated from 45% of the disinfected clips under anaerobic growth conditions. Strains of Staphylococcus, Streptococcus, Prevotella, Nisseria, Kocuria, Actinomyces, Selenomonas, Neisseria, and Bacillus were sampled from the surfaces of single post-disinfection clips in aerobic or anaerobic growth conditions less frequently. The Streptococci strains are mainly found among the normal oral flora and upper respiratory tract.11 Accumulated data also demonstrate the association of anaerobic strains of Prophyromonas, Prevotella, Fusobacterium, Selenomonas, and Eubacterium in chronic or refractory periodontitis.7-13 These bacteria can also be found in individuals with good periodontal health in which these bacteria co-exist harmlessly with other oral flora in the host.8,13 Due to the nature of the treatment provided at the hygiene clinic, finding the presence of oral bacteria involved in periodontitis, such as Treponema socranskii, Tannerella forsythensis, and Parvimonas micra, on the clips immediately after treatment was expected.8,10-12 However, in this study, none of the oral bacteria species found in samples on post-disinfection clips were considered periodontal pathogens.
The present study at the hygiene clinic revealed that after treatment and in addition to environmental bacteria, skin and oral bacteria can be found on the surfaces of clips. After disinfection, the bacterial species that survived on the clips were predominantly skin isolates; albeit in a low number, oral species of bacteria were also found on individual clips. How did these bacteria accrue on the surfaces of these clips? Common and predominant skin bacteria, such as S epidermidis and P acnes, could be transferred from the hands of dental staff members to the clips prior to treatment sessions. These bacteria could also be transferred from the patient’s hands if the patient touched the clips. A patient’s saliva and the spray or spatter produced during the hygiene treatment may contribute to the presence of oral bacteria on the clips’ surfaces. In addition, oral bacteria could be inadvertently transferred onto the clips from the gloved hands of the dentist, hygienist, and/or assistant during treatment performed in the patient’s oral cavity. Finally, environmental and airborne bacteria, such as strains of Bacillus, Micrococcus, and Pseudomonas, can survive in waterlines and the clinic’s air1 and contaminate the clips. From an infection control perspective, bacteria and viruses can be transmitted through contact with patient materials, contaminated equipment and instruments, spatter generated during treatment, and inhalation of environmental and airborne microorganisms.1 However, to cause a chain of infection, suitable numbers of virulent pathogenic microorganisms and the materials containing contaminants, a transmission mode from the source to the host, and a suitable portal of entry into a vulnerable host should all occur simultaneously. By preventing at least one of these parameters, potential spread of diseases can be prevented.1 The authors’ study at the hygiene clinic demonstrated that with the current disinfection protocol, specific aerobic, as well as anaerobic bacteria, can remain viable on the surfaces of clips after disinfection, recover, and grow on a supportive nutrient agar and optimal growth conditions. In the settings of this study, the finding that bacterial contaminants can persist on the surfaces of disinfected clips has not been clinically shown to be the cause of an infectious disease problem and actual transmission to patients was not demonstrated. However, some of the ubiquitous bacteria found may potentially become opportunistic pathogens in appropriate physical conditions, such as in predisposed patients or clinicians.
Conclusions
The results showed that after hygiene treatment and, to a significantly lesser extent, after disinfection, various environmental, skin, and oral bacteria can be isolated from the surfaces of reusable bib clips. While the disinfection procedure is significantly effective, 40% and 70% of the tested clips still harbored one or more aerobic and anaerobic bacteria, respectively; none of these bacteria was considered to be periodontal pathogens. Clinically, the mere presence of bacteria on disinfected clips has not been shown to trigger infectious disease problems. However, the findings suggest that the nature of the treatment presented at specific clinics may affect the bacterial loads found on the clips and the composition of those bacterial loads. This indicates that each dental clinic may face different cross-contamination risks.
Disclosure
The authors have no conflict of interest. The study was sponsored in part by Dux Dental (www.duxdental.com).
Acknowledgments
The authors would like to thank the staff of the Department of Periodontology at Tufts School of Dental Medicine for being welcoming and supportive during the study. They would also like to thank Shannon Belletto, who is the infection control administrator at TUSDM, for her helpful discussion and comments. Finally, they thank the staff members of the Human Microbial Identification Microarray Core at the Forsyth Institute for their help in this study.
References
1. Kohn WG, Collins AS, Cleveland JL, et al, Centers for Disease Control and Prevention (CDC). Guidelines for infection control in dental health-care settings—2003. MMWR Recomm Rep. 2003;52(RR-17):1-61.
2. Molinari JA. Microbial contamination of patient napkin holders. The Dental Advisor. 2010;29:1-3.
3. Study: Bacteria found on 70% of dental bib holders [news release]. San Francisco, CA: DrBicuspid.com; March 19, 2012. https://www.drbicuspid.com/index.aspx?sec=sup&sub=hyg&pag=dis&ItemID=310087. Accessed February 19, 2013.
4. Study finds bib chain potential source of bacteria. Dental Health Magazine; March 16, 2010. https://worldental.org/dental-news/study-finds-bib-chain-potential-source-of-bacteria/1184. Accessed February 19, 2013.
5. Alt-Holland A, Srinivasan S, Lucier R, et al. Do bib clips pose a cross-contamination risk at the dental clinic? Compend Contin Educ Dent. 2012;33(9 suppl):1-7.
6. Alt-Holland A, Thondapu RS, Rajapaksha KS, et al. A novel method to examine bacterial contamination in dental clinics [abstract]. J Dent Res. 2012;91(spec iss A). Abstract 659.
7. Faveri M, Mayer MP, Feres M, et al. Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol Immunol. 2008;23(2):112-118.
8. Colombo AP, Boches SK, Cotton SL, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80(9):1421-1432.
9. Paster BJ, Dewhirst FE. Molecular microbial diagnosis. Periodontol 2000. 2009;51:38-44.
10. Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42(1):80-87.
11. Aas JA, Paster BJ, Stokes LN, et al. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43(11):5721-5732.
12. Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002-5017.
13. Mane AK, Karmarkar AP, Bharadwaj RS. Anaerobic bacteria in subjects with chronic periodontitis and in periodontal health. J Oral Health Comm Dent. 2009;3(3):49-51.
Related Content:
The authors’ previous study, “Do Bib Clips Pose a Cross-Contamination Risk at the Dental Clinic?,” was published as a supplement to Compendium of Continuing Education in Dentistry in October 2012. It is available online at: www.dentalaegis.com/special-issues/Dux/2012/08
About The Authors
Addy Alt-Holland, BSc, MSc, PhD
Department of Endodontics
Tufts University School of Dental Medicine
Boston, Massachusetts
Christina M. Murphy, BS
Department of Microbiology
The Forsyth Institute
Cambridge, Massachusetts
Anne Powers, RDH
Department of Periodontology
Tufts University School of Dental Medicine
Boston, Massachusetts
Claire L. Kublin, BS
Department of Research Administration
Tufts University School of Dental Medicine
Boston, Massachusetts
Youjin Natalie Jeong, DMD
Department of Periodontology
Tufts University School of Dental Medicine
Boston, Massachusetts
Michelle DiMattia
Mount Ida College for Dental Hygiene
Newton, Massachusetts
Linh Pham
Mount Ida College for Dental Hygiene
Newton, Massachusetts
Angel Park, MS, MPH
Department of Research Administration
Tufts University School of Dental Medicine
Boston, Massachusetts
Matthew Finkelman, PhD
Department of Public Health and Community Service
Tufts University School of Dental Medicine
Boston, Massachusetts
Maureen Lombard, RDH, MM
Department of Clinical Affair
Tufts University School of Dental Medicin
Boston, Massachusetts
James B. Hanley, DMD
Department of Periodontology and Department of Clinical Affairs
Tufts University School of Dental Medicine
Boston, Massachusetts
Bruce J. Paster, PhD
Department of Microbiology
The Forsyth Institute,
Cambridge, Massachusetts
Department of Oral Medicine
Infection & Immunity
Harvard School of Dental Medicine
Boston, Massachusetts
Gerard Kugel, DMD, PhD
Department of Research Administration
Tufts University School of Dental Medicine
Boston, Massachusetts